Research Highlights
Training AI with synthetic data for measuring crop seed shape Development of an efficient method to accelerate the development of machine learning models for plant phenotyping.
A team of scientists led by Yosuke Toda, Designated Assistant Professor at the Institute of Transformative Bio-Molecules (WPI-ITbM), Nagoya University, and Fumio Okura, Assistant Professor at the Institute of Scientific and Industrial Research,Osaka University, have developed a system which utilizes image analysis and artificial intelligence (AI) to automatically and precisely analyze the shape of large numbersof seeds from a single image. As the shape of the seed is an important agronomic trait that is closely linked to the yield and quality of crops, a method for automatically determining and evaluating suchfrom an image is an indispensable tool for plant breeding. However, creating the training dataset is laborious and time consuming, especially when the number of objects to annotate is as large as it is in the case of seeds. To date,it has been difficult to quickly and conveniently analyze the number of seeds of different crop species at once.
About the research:
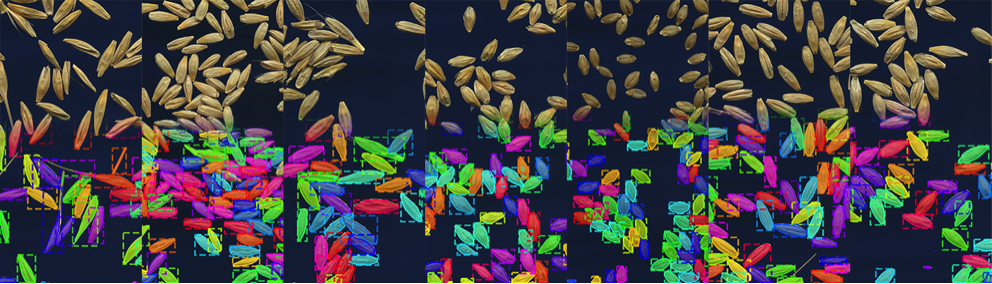
In the midst of unprecedented climate change and population growth, establishment of a method to rapidly create elite crop varieties via selective breeding is a matter of urgency to maintain the food supply. In order to select such cultivars, it is necessary to define and evaluate a metric for what is a 'superior variety' in an efficient manner. For example, the shape of seeds(their length and diameter, for example) is understood to be a trait (i.e. phenotype) closely linked to the quality and yield of crops, and is thus an important factor when conducting selective breeding. However, as measuring the characteristics of each tiny seed one by one by hand is laborious and time consuming, there is a great desire for automation using machine learning methods. In recent years, image analysis using deep learning has entered active use in the field of plant phenotyping. In this study, using instance segmentation based on deep learning, the research team aimed to develop a system that can automatically acquire data on hundreds of seeds from just one image (Figure 1).
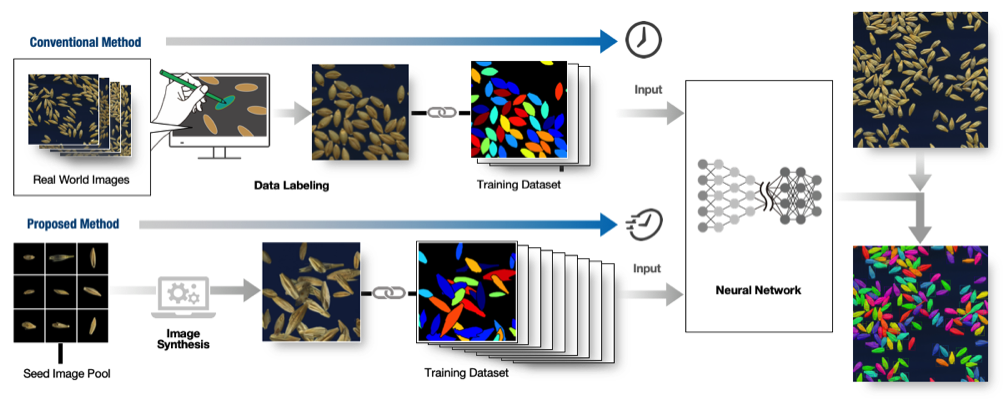
Training data is required to make use of (supervised) deep learning. Usually, training data for instance segmentation is prepared by hand, for example by labeling every object in the images with different colors. However, for examples such as the seeds in Figure 1, whose number is vast, creating the training data is very time consuming (for example, having to individually color hundreds of seeds for tens or hundreds of images for each seed variety). Thus, it has been considered difficult to generate a machine learning model that can quickly and simply analyze the seed shapes of different varieties or species (Figure 2, above).
The Proposed Method
Dr. Toda's research group succeeded in creating a large volume of training data from only a small number of seeds to effectively train the machine learning (deep learning) model. This method is one example of an approach calleddomain randomization, and spares the effort involved in creating the training data, accelerating the development of machine learning models. In the proposed method, sample images of a small number of barley seeds whose shape information was already know (cf. the seed image pool in Figure 2) were randomly arranged in virtual space, creating a large number and variety of synthesized images (Figure 2, below). The model trained with this datasetwas able to detect the seeds and extract their shape data with the same degree of accuracy as when done by hand. Notably, no hand annotated training dataset was required.
This system is able to handle various arrangements of seeds, and even pick out and analyze individual seeds when simply sprinkled at random as in Figure 1. Their detailed characteristics, beyond just length and diameter, can be drawn from the shape data acquired through instance segmentation. The experiment actually highlighted that the system can clearly identify the characteristic differences in shape of each crop. It is expected that in the future it will be possible to measure fine differences in the growth environment and variety, becoming a powerful tool for plant breeding.
Furthermore, the study showed that the same method can readily be employed to measure the seeds of a variety of different crops, such as rice, wheat, oats and lettuce. These results strongly suggest that, regardless of crop, it is possible to make the automatic measurement of large numbers of seeds a reality.
Future development
This study has realized the simple and efficient shape analysis of crop seeds. To ensure an uninterrupted food supply in a time of extensive climate change, it is crucial to speed up the cycle of selective breeding and rapidly create superior varieties. Variety evaluation measures employing seed shape will be a strong foundation tool for selective breeding. Additionally, beyond variety evaluation, it is expected to contribute to the plant science domain by revealing characteristics of seeds not formerly observed by the human eye.
The majority of research into instance segmentation-based image analysis is conducted using existing datasets including things such as people and cars. On the other hand, plant image analysis has a variety of its own characteristics. Since there is great variation in plants' species, location and individual appearance, different training data is needed for respective applications. While this is also the case for others with multiple applications, the creation of new training data for plants is particularly difficult. The method of generating synthetic training data employed in this study can be used in a variety of applications. Based on the initiative of this research, it is expected that it will be possible to go beyond the analysis of seeds, and accelerate the development of a machine learning model for the measurement of plant phenotypes.
Vocabulary
1) Plant phenotyping
A number of plant varieties are created and selected in the breeding of plants. When doing so, the ability to be able to assess what is a 'good variety' in a quick and efficient manner is important. Plant phenotyping is a process that employs the qualitative and quantitative measuring of plants' appearance (their phenotype), making it possible to evaluate whether they are a good or bad variety. The targets for measurement are not only the seeds, but also the above-ground sections of the plant (the stems, leaves and fruit, for example), the roots and the functions such as photosynthesis and desiccation resistance. Image analysis-based plant phenotyping, which allows for the efficient collection of detailed information about a large number of individuals, has been the subject of enthusiastic research in recent years.
2) Instance segmentation
A method for detecting and segmenting individual objects found in an image one by one. Recently, instance segmentation by deep learning has been being actively developed and applied across a variety of fields. For example, this technology is not only expected to find use in the onboard imaging systems of self-driving cars for detecting other cars and people, it is also being used in the plant phenotyping field for the detection and regional division of leaves. The issue with its practical use is the effort required to create training data, which amounts to at least tens of images of regionally divided objects.
3) Domain randomization
When generating the training dataset for deep learning by computer simulation, differences between those synthetic images and the real environment are often problematic. Domain randomization is a method that, by randomly replicating various environmental differences during the image creation process, aims to make up for this gap without needing to produce data in the real environment. In the past, various different randomizations have been tried, such as light sources and object locations. In this study, by arranging the images of just a few seeds at random location and orientation, it was possible to cover a variety of seed configurations found in the real world.
Journal Information:
The article "Training instance segmentation neural network with synthetic datasets for crop seed phenotyping" by Yosuke Toda, Fumio Okura, Jun Ito, Satoshi Okada, Toshinori Kinoshita, Hiroyuki Tsuji and Daisuke Saisho is published in Nature Communications Biology
DOI: 10.1038/s42003-020-0905-5I
2020-04-16