
Teruki Honma, PhD
Team Leader, Laboratory for Structure-Based Molecular Design,
RIKEN Center for Biosystems Dynamics Research
Lecture Title
Development of drug discovery AI platform combining prediction AI and generative AI (AMED DAIIA)
Abstract
Since the latter half of the 2010s, the development of artificial intelligence (AI) methods including deep learning, the lowering of the hurdles to obtain big data, and the development of hardware that can be used for AI such as general-purpose GPUs have led to AI has become more useful and practical. In Japan, in 2016, led by Prof. Okuno (Kyoto University), Life Intelligence Consortium (LINC) was launched to accelerate the applications of AI to the entire life science fields, including drug discovery and medical care. Through the 1st phase of LINC, we have developed various AI-related elemental technologies by building and verifying prototypes of AI that can be used in various fields, mainly using public data. In addition, mutual understanding and sharing of future issues among life-related companies, IT companies, and academic research institutes progressed.
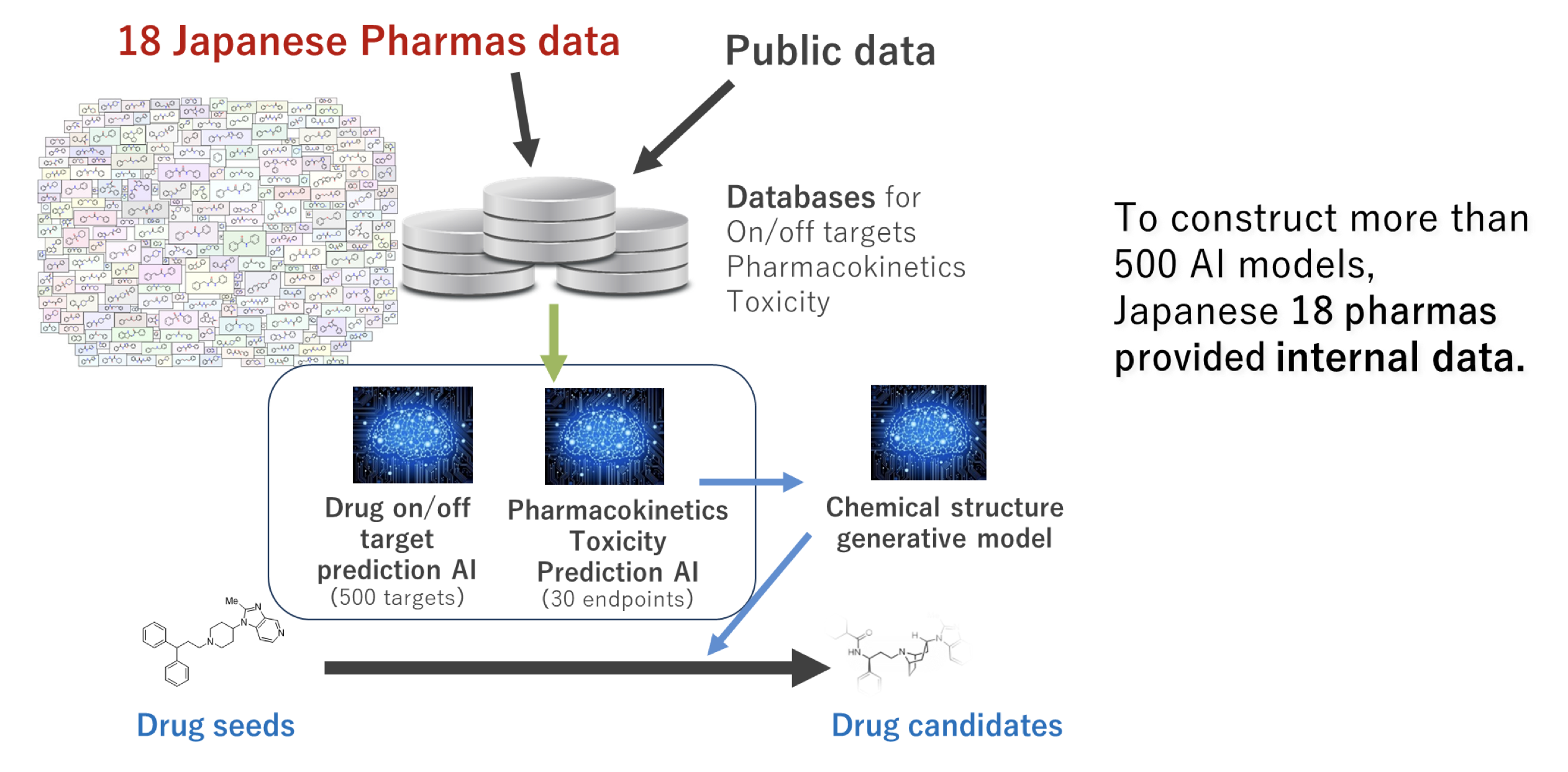
In the next phase of social implementation of LINC, it is very important from the viewpoint of practicality to develop AI models by integrating not only public data but also confidential data historically collected in companies. Under such circumstances, a project of the Japan Agency for Medical Research and Development (AMED) named “DAIIA” to build a drug discovery AI platform by linking AI technologies of academia and drug discovery-related assay data of pharmaceutical companies started in August 2020.
The AMED DAIIA project has built an AI platform that comprehensively supports drug design from hits to development candidates by combining AI models that comprehensively predict compound profiles such as on/off targets and ADMET, and AI that generates new structures (generative model). The goal is to streamline drug discovery for small and middle-sized molecules. In particular, we are focusing on off-targets, building prediction models for more than several hundred targets, and measuring new data for 44 targets that are particularly important for side effects. As for learning data, in addition to public data, we are planning to receive in-house data and learning results from 18 pharmaceutical companies. In terms of key technologies, we are developing and implementing multimodal learning by GCN, semi-supervised/self-supervised learning, multi-objective optimization by reinforcement learning, and federated learning. In particular, we are promoting development with an emphasis on practicality in the field of drug discovery and are working on fine tuning of AI models using new internal data in ongoing projects, visualization of the applicability domain, and use for structure generation.
Laboratory for Structure-Based Molecular Design website
https://www.bdr.riken.jp/en/research/labs/honma-t/index.html