The 4th Tsuneko & Reiji Okazaki Award 2018
Cyril Zipfel (University of Zurich, Switzerland)

Maria Barna
Assistant Professor, Department of Developmental Biology and Genetics, Stanford University School of
Medicine
The central dogma of molecular biology has for decades served as an explanation for the flow of genetic information within a biological system. In so far as the transmission of biological information, the ribosome has been perceived to decode the genome with machine-like precision; serving as an integral but largely passive participant in the synthesis of all proteins across all kingdoms of life. Our research has fundamentally changed this view, by demonstrating that not all of the millions of ribosomes within each cell are the same and that ribosome heterogeneity provides a novel means for diversity of the proteins that can be produced in specific cells, tissues, and organisms. Collectively, we have termed this additional layer of gene regulation as a “ribocode”, which enables ribosomes to be tuned to translate specific mRNAs. This talk will highlight our research effort to apply sophisticated mass spectrometry, computational biology, genomics and developmental biology to characterize the impact of ribosome heterogeneity on translation of the genetic code in time and space. We have identified specific classes of ribosomes that are essential for forming the mammalian body plan and others that are likely to control cell signaling, metabolism, proliferation, cell death, and embryonic development. We have also discovered the mammalian ribo-interactome: a compendium of hundreds of new ribosome associated proteins (RAPs) that further diversify ribosome specificity at key subcellular organelles, such as the endoplasmic reticulum. I will further present recent findings on the mechanisms by which ribosome-mediated control of gene expression is encoded by structured RNA regulons within 5’UTRs. Together, these studies reveal a critical link between ribosome heterogeneity and specialized translational control of the mammalian genome, which adds an important layer of control to gene expression.
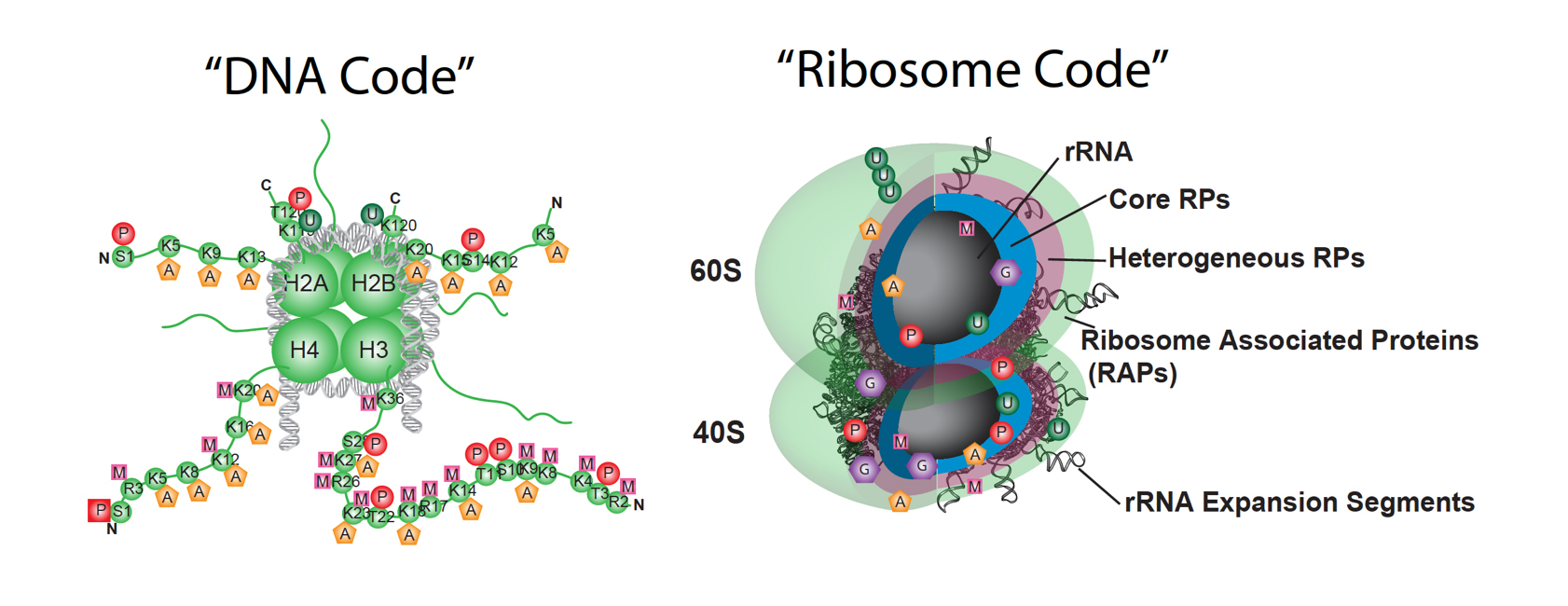
Figure: Parallels between a DNA and ribosome code. The eukaryotic ribosome is a molecular machine composed of 4 RNA molecules and 79 unique ribosomal proteins (RPs) depicted as colored ovals. Several layers of regulation could confer more specialized ribosome function. For example, similar to DNA, rRNA is extensively modified. Changes in RP modifications and/or composition may confer greater specificity to the RNA-based translation machinery, as do histone modifications to chromatin.
Reference
| Date | Monday, November 20th, 2017 |
|---|---|
| Time | 16:35 - 17:50 |
| Place | Noyori Conference Hall, Nagoya University |
| Website | https://www.itbm.nagoya-u.ac.jp/istbm-5/program_jp.html#okazaki |