Research Highlights
A new method for in vivo plant cell imaging with SNAP-tag proteins
The Nagoya University Institute of Transformative Bio-Molecules (WPI-ITbM) research team of postdoctoral fellows Ryu Iwatate, Akira Yoshinari and Noriyoshi Yagi, Professor Wolf Frommer and Designated Lecturer Masayoshi Nakamura, in collaboration with Associate Professor Mako Kamiya and Designated Assistant Professor Toru Komatsu of the University of Tokyo and the research group of ITbM's Professor Shigehiro Yamaguchi, have succeeded in visualizing the microtubule dynamics and cell membrane protein endocytosis 1) of living plant cells via a method using SNAP-tag 2) and a synthetic fluorescent probe.
This discovery, providing a new method for fluorescent marking of proteins in plant cells, represents an important advancement in plant cell biology. It is expected to find use in super-resolution imaging using extremely light stable markers, and techniques for determining place- and time-specific pH and Ca2+ levels.
The results of this study were published in the online edition of the American journal The Plant Cell on 5 August 2020.
About the research
The marking of specific proteins in live specimens by synthetic dyes has garnered a great deal of attention recently in the field of fluorescent imaging. The method using SNAP-tag is one of these - by using a synthetic dye connected to specific ligands such as benzylguanine derivatives, it is possible to bind the dye to the desired protein fused to the SNAP-tag. SNAP-tag visualization of in vivo protein dynamics makes a wide range of contributions to medical and biological study, for example in cancer research. This method has a number of advantages, including its ability to work with a wide range of dyeing compounds and to mark the targeted protein at a specified time. However, live cell imaging 3) using SNAP-tag and synthetic dyes to mark proteins is not being performed in the field of plant science, as synthetic dyes are unable to reach the target proteins due to the presence of the cell wall in plant cells.
This time, the research team demonstrated that it was possible to perform live cell imaging using SNAP-tag even in plants using three types of synthetic dyes which bond to the SNAP-tag to mark microtubules, part of the cytoskeleton. Furthermore, by using the synthetic dye DRBG-488, which does not permeate the cell membrane and fluoresces only when it bonds with SNAP-tag, they were able to exclusively mark the auxin transporters in the cell membrane and succeeded in visualizing the process of membrane proteins being taken into the cell (endocytosis) after they had been marked. Thus, they were able to clearly differentiate between transporter proteins which had been newly synthesized inside the cell and those that had been taken into the cell by endocytosis. At the same time, using tobacco BY-2 cells, they investigated and listed which out of 31 dyes were able to enter the cell. Interestingly, it was found that 23 synthetic dyes were taken into the BY-2 cells, that most of the synthetic dyes could be used with SNAP-tag to mark cytosolic components in plant cells, and that those which could not permeate the cell membrane could be used in marking membrane proteins from outside the cell.
The findings of this study provide a new technique for the fluorescent marking of proteins in plant cells, and represent an important step forward in plant cell biology research. It is expected to find use in super-resolution imaging using extremely light stable markers, and techniques for determining place- and time-specific pH and Ca2+levels.
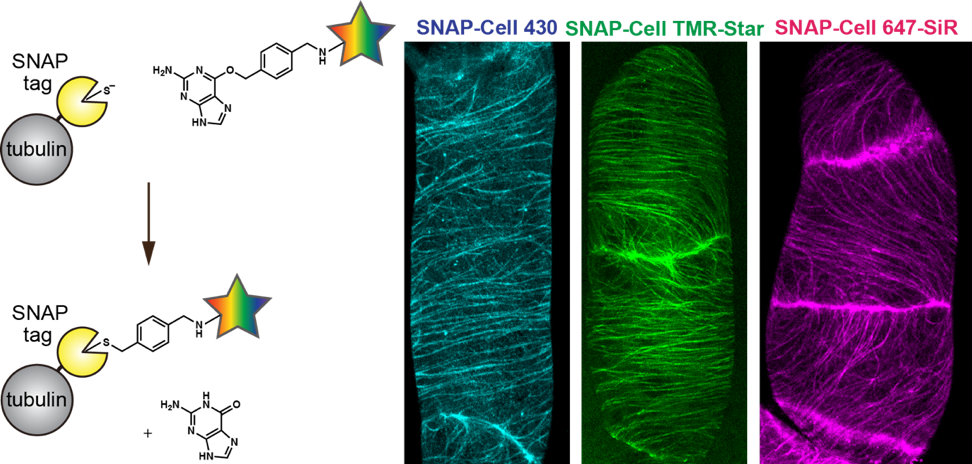
Reaction of fluorescent substrate with SNAP-tag bound to tubulin protein (left), microtubules in BY-2 cells marked with three different synthetic dyes (right)
Vocabulary
1) Endocytosis
The process by which proteins in the cell membrane and materials outside the cell are taken up by cell membrane invagination and the formation of the endoplasmic reticulum.
2) SNAP-tag
An enzyme tag of around 20kDa consisting of genetically engineered human O6-alkylguanine-DNA alkyltransferase (hTAG). Forms covalent bonds with substrates containing benzyl guanine and chlorobenzyl pyrimidine groups. Using SNAP-tag, it is possible to add a fluorescent dye to a target protein
3) Live cell imaging
The observation in living cells of proteins and organelles which have been marked with, for example, a fluorescent probe.
Journal information
The article "Covalent self-labeling of tagged proteins with chemical fluorescent dyes in BY-2 cells and Arabidopsis seedlings" by Ryu J. Iwatate, Akira Yoshinari, Noriyoshi Yagi, Marek Grzybowski, Hiroaki Ogasawara, Mako Kamiya, Toru Komatsu, Masayasu Taki, Shigehiro Yamaguchi, Wolf B. Frommer and Masayoshi Nakamura is published in The Plant Cell
DOI: https://doi.org/10.1105/tpc.20.00439
2020-08-07

